Differentiable Electron Microscopy Simulation: Methods and Applications for Visualization
Abstract
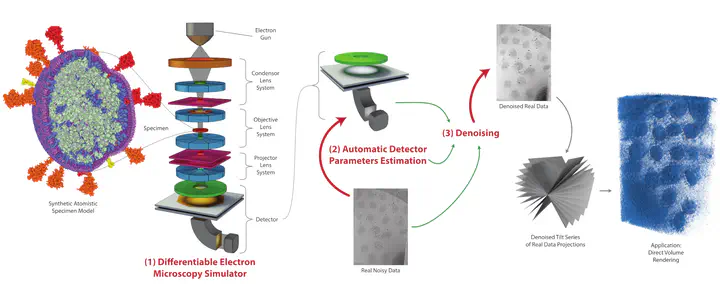
We propose a new microscopy simulation system that can depict atomistic models in a micrograph visual style, similar to results of physical electron microscopy imaging. This system is scalable, able to represent simulation of electron microscopy of tens of viral particles and synthesizes the image faster than previous methods. On top of that, the simulator is differentiable, both its deterministic as well as stochastic stages that form signal and noise representations in the micrograph. This notable property has the capability for solving inverse problems by means of optimization and thus allows for generation of microscopy simulations using the parameter settings estimated from real data. We demonstrate this learning capability through two applications: (1) estimating the parameters of the modulation transfer function defining the detector properties of the simulated and real micrographs, and (2) denoising the real data based on parameters trained from the simulated examples. While current simulators do not support any parameter estimation due to their forward design, we show that the results obtained using estimated parameters are very similar to the results of real micrographs. Additionally, we evaluate the denoising capabilities of our approach and show that the results showed an improvement over state-of-the-art methods. Denoised micrographs exhibit less noise in the tilt-series tomography reconstructions, ultimately reducing the visual dominance of noise in direct volume rendering of microscopy tomograms.